Keywords
dipole moment variation upon excitation.
fluorescent proteins
machine learning
QM/MM molecular dynamics
Abstract
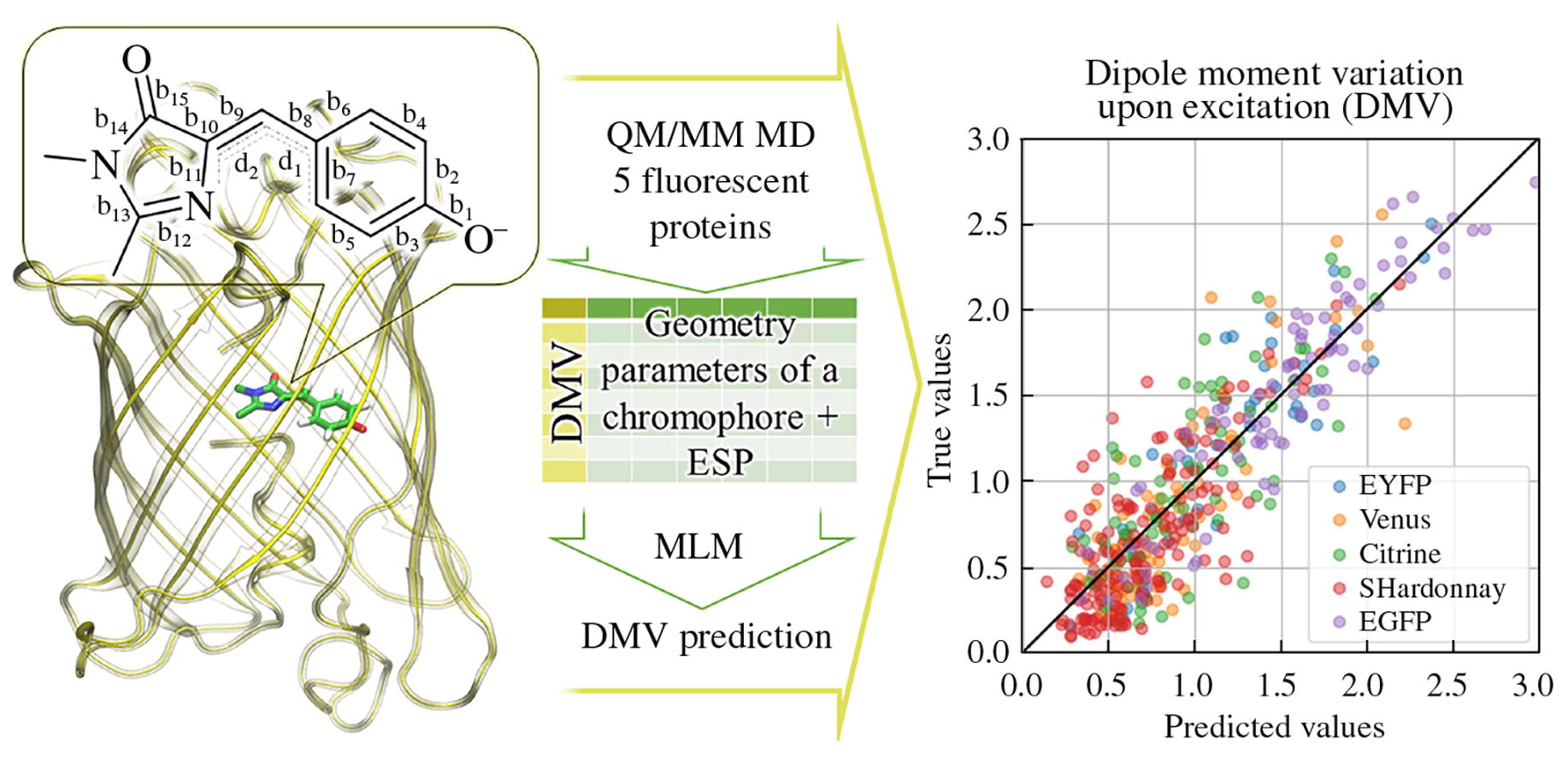
We demonstrate that machine learning models trained on a set of features obtained from QM/MM molecular dynamic trajectories of fluorescent proteins can be used to predict the chromophore dipole moment variation upon excitation, the quantity related to the electronic excitation energy. Linear regression, gradient boosting, and artificial neural network- based models were considered using cross-validation on the training dataset. Gradient boosting approach proved to be the most accurate for both internal (R2 = 0.77) and external (R2 = 0.7) test sets.
References
.

Adamo C., Barone V.
Journal of Chemical Physics,
1999
.

Grimme S., Antony J., Ehrlich S., Krieg H.
Journal of Chemical Physics,
2010
.

Best R.B., Zhu X., Shim J., Lopes P.E., Mittal J., Feig M., MacKerell A.D.
Journal of Chemical Theory and Computation,
2012
.

Word J.M., Lovell S.C., Richardson J.S., Richardson D.C.
Journal of Molecular Biology,
1999
.

Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L.
Journal of Chemical Physics,
1983
.

Melo M.C., Bernardi R.C., Rudack T., Scheurer M., Riplinger C., Phillips J.C., Maia J.D., Rocha G.B., Ribeiro J.V., Stone J.E., Neese F., Schulten K., Luthey-Schulten Z.
Nature Methods,
2018
.

Phillips J.C., Hardy D.J., Maia J.D., Stone J.E., Ribeiro J.V., Bernardi R.C., Buch R., Fiorin G., Hénin J., Jiang W., McGreevy R., Melo M.C., Radak B.K., Skeel R.D., Singharoy A., et. al.
Journal of Chemical Physics,
2020
.

Khrenova M.G., Mulashkin F.D., Bulavko E.S., Zakharova T.M., Nemukhin A.V.
Journal of Chemical Information and Modeling,
2020
.

Acharya A., Bogdanov A.M., Grigorenko B.L., Bravaya K.B., Nemukhin A.V., Lukyanov K.A., Krylov A.I.
Chemical Reviews,
2016
.

Lin C., Boxer S.G.
Journal of the American Chemical Society,
2020
.

Drobizhev M., Tillo S., Makarov N.S., Hughes T.E., Rebane A.
Journal of Physical Chemistry B,
2009
.

Lin C., Romei M.G., Oltrogge L.M., Mathews I.I., Boxer S.G.
Journal of the American Chemical Society,
2019
.

Drobizhev M., Callis P.R., Nifosì R., Wicks G., Stoltzfus C., Barnett L., Hughes T.E., Sullivan P., Rebane A.
Scientific Reports,
2015
.
Nifosì R., Mennucci B., Filippi C.
Physical Chemistry Chemical Physics,
2019
.

Day R.N., Davidson M.W.
Chemical Society Reviews,
2009
.

Griesbeck O., Baird G.S., Campbell R.E., Zacharias D.A., Tsien R.Y.
Journal of Biological Chemistry,
2001
.

Chai J., Head-Gordon M.
Physical Chemistry Chemical Physics,
2008
.

Lippincott-Schwartz J., Patterson G.H.
Trends in Cell Biology,
2009
.

Denning E.J., Priyakumar U.D., Nilsson L., Mackerell A.D.
Journal of Computational Chemistry,
2011
.

Rekas A., Alattia J., Nagai T., Miyawaki A., Ikura M.
Journal of Biological Chemistry,
2002
.

Shinoda H., Shannon M., Nagai T.
International Journal of Molecular Sciences,
2018
.

Improving the Second-Order Nonlinear Optical Response of Fluorescent Proteins: The Symmetry Argument
De Meulenaere E., Nguyen Bich N., de Wergifosse M., Van Hecke K., Van Meervelt L., Vanderleyden J., Champagne B., Clays K.
Journal of the American Chemical Society,
2013
.

Saito Y., Oikawa M., Nakazawa H., Niide T., Kameda T., Tsuda K., Umetsu M.
ACS Synthetic Biology,
2018
.

Tam C., Zhang K.Y.
Proteins: Structure, Function and Genetics,
2021
.

Arpino J.A., Rizkallah P.J., Jones D.D.
PLoS ONE,
2012
.

Gonzalez Somermeyer L., Fleiss A., Mishin A.S., Bozhanova N.G., Igolkina A.A., Meiler J., Alaball Pujol M., Putintseva E.V., Sarkisyan K.S., Kondrashov F.A.
eLife,
2022
.

Yang K.K., Wu Z., Arnold F.H.
Nature Methods,
2019
.

Wittmann B.J., Johnston K.E., Wu Z., Arnold F.H.
Current Opinion in Structural Biology,
2021
.

Willig K.I., Wegner W., Müller A., Clavet-Fournier V., Steffens H.
Cell Reports,
2021
.

Li M., Kang L., Xiong Y., Wang Y.G., Fan G., Tan P., Hong L.
Journal of Cheminformatics,
2023