Keywords
ATP hydrolysis
Laplacian of electron density.
machine learning
myosin
QM/MM molecular dynamics
Abstract
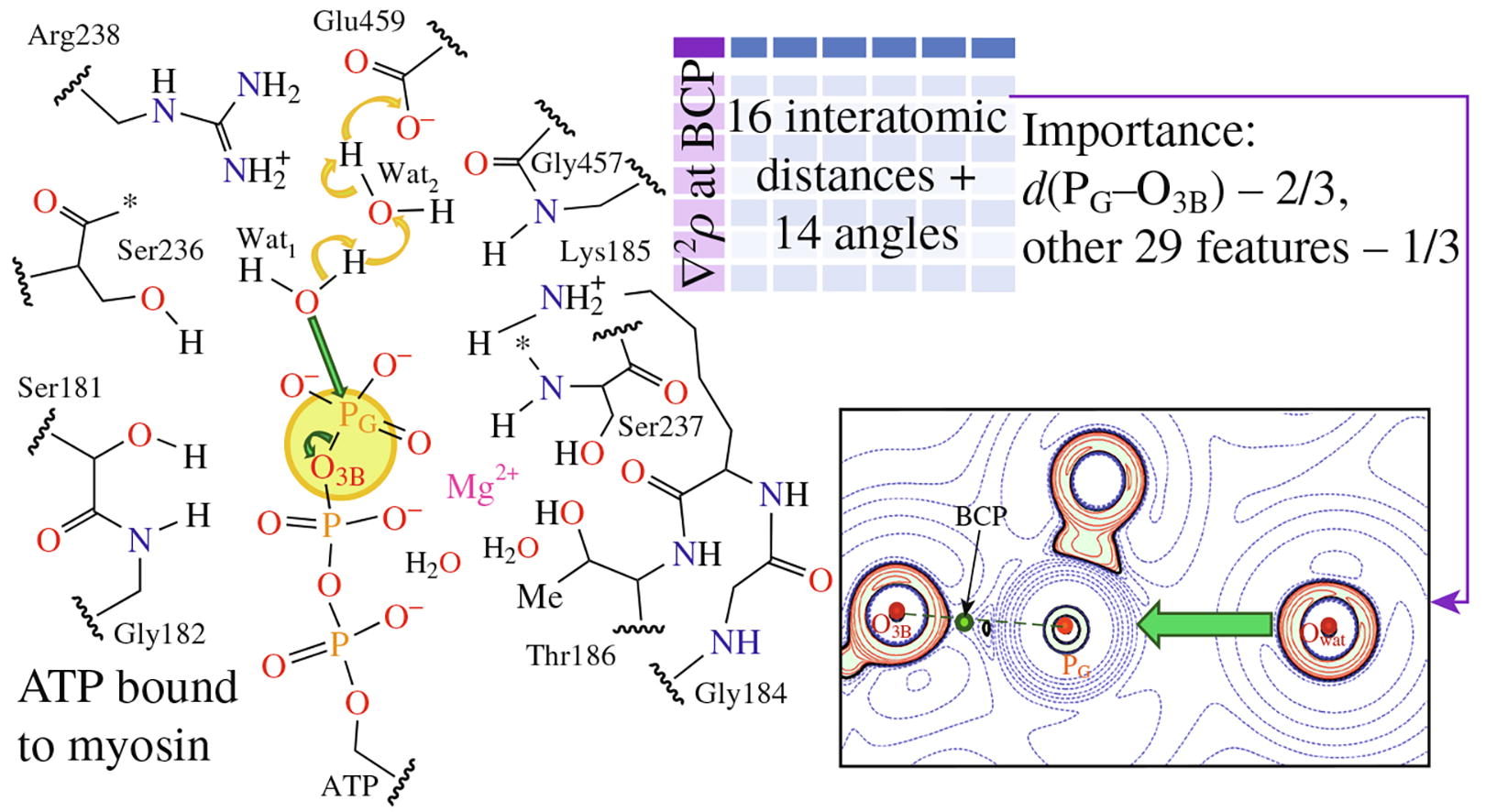
Molecular dynamic simulations using QM/MM potentials are performed for the enzyme-substrate complex of adenosine triphosphate (ATP) with the motor protein myosin. Machine learning methods are applied to a dataset consisting of the geometry parameters of the active site in the enzyme-substrate complex to predict the Laplacian of electron density at the bond critical point of the PG-O3B bond being broken in ATP. Using a gradient boosting machine learning model, a mean absolute error of 0.01 a.u. and an R2 score of 0.99 are achieved, and it is found that the PG-O3B bond length is the most important feature, contributing 2/3, while other geometry features contribute 1/3.
References
.

Humphrey W., Dalke A., Schulten K.
Journal of Molecular Graphics,
1996
.

Lu T., Chen F.
Journal of Computational Chemistry,
2011
.

Adamo C., Barone V.
Journal of Chemical Physics,
1999
.

Grimme S., Antony J., Ehrlich S., Krieg H.
Journal of Chemical Physics,
2010
.

Khrenova M.G., Grigorenko B.L., Nemukhin A.V.
ACS Catalysis,
2021
.

Smith C.A., Rayment I.
Biochemistry,
1996
.
Gribanova T.N., Minyaev R.M., Minkin V.I.
Mendeleev Communications,
2023
.

Best R.B., Zhu X., Shim J., Lopes P.E., Mittal J., Feig M., MacKerell A.D.
Journal of Chemical Theory and Computation,
2012
.

Word J.M., Lovell S.C., Richardson J.S., Richardson D.C.
Journal of Molecular Biology,
1999
.

Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L.
Journal of Chemical Physics,
1983
.

Martyna G.J., Klein M.L., Tuckerman M.
Journal of Chemical Physics,
1992
.

Melo M.C., Bernardi R.C., Rudack T., Scheurer M., Riplinger C., Phillips J.C., Maia J.D., Rocha G.B., Ribeiro J.V., Stone J.E., Neese F., Schulten K., Luthey-Schulten Z.
Nature Methods,
2018
.

Phillips J.C., Hardy D.J., Maia J.D., Stone J.E., Ribeiro J.V., Bernardi R.C., Buch R., Fiorin G., Hénin J., Jiang W., McGreevy R., Melo M.C., Radak B.K., Skeel R.D., Singharoy A., et. al.
Journal of Chemical Physics,
2020
.

Lundberg S.M., Erion G., Chen H., DeGrave A., Prutkin J.M., Nair B., Katz R., Himmelfarb J., Bansal N., Lee S.
Nature Machine Intelligence,
2020
.

Singer K., Smith W.
Molecular Physics,
1988
.

Hartman M.A., Spudich J.A.
Journal of Cell Science,
2012
.
Rumyantcev R.V., Yu. Zhigulin G., Zabrodina G.S., Katkova M.A., Yu. Ketkov S., Fukin G.K.
Mendeleev Communications,
2023
.
Khrenova M.G., Mulashkina T.I., Stepanyuk R.A., Nemukhin A.V.
Mendeleev Communications,
2024
.

Chakraborti A., Tardiff J.C., Schwartz S.D.
Journal of Physical Chemistry B,
2024
.
Khrenova M.G., Mulashkina T.I., Kulakova A.M., Polyakov I.V., Nemukhin A.V.
Moscow University Chemistry Bulletin,
2024